closed
Luigina De Leo, IRCCS Materno Infantile Burlo Garofolo, Trieste, Italy
3-years Project
Celiac disease
Area: Genetics
Grant: 003/2020
- Title: DNA methylation and transcriptional profiles of intestinal epithelium from celiac patients
- Duration: 3-years Project
- Principal Investigator: Luigina De Leo, IRCCS Burlo Garofolo, Trieste, Italy
- Publications: manuscript in preparation
THE STUDY
Project rational and aims
Epigenetics is a highly plausible mechanism that may both initiate and maintain intestinal mucosal inflammation in several immune diseases, including inflammatory bowel disease, type 1 diabetes, and different cancers. The importance of studying individual disease-relevant cell types to best identify molecular alterations involved in pathophysiology has been demonstrated. Therefore, the analysis of highly purified intestinal epithelial cells (IECs), bearing the epithelial cell adhesion molecule CD326, could provide relevant information.
We hypothesized that an epigenetic and transcriptomic profiling of IECs from children newly diagnosed with classical or potential Celiac Disease (CD), compared with controls, could reveal specific disease-associated alterations to learn more about the pathogenesis of CD in all its clinical and biological manifestations.
Our aims were:
- Enrolling patients with classical and potential CD at diagnosis, gluten-free diet (GFD)-treated CD patients, and a matched cohort of controls suffering from other gastrointestinal disorders.
2.Profiling the genome-wide DNA methylation and gene expression of IECs derived from the enrolled patients to detect genes differentially methylated and expressed in the three study groups (classical CD vs controls, potential CD vs controls, and GFD-treated CD patients vs controls).
3.Generating intestinal epithelial organoids from patients’ biopsy samples to investigate if disease-associated epigenetic alterations are retained in ex-vivo organoid cultures.
4.Since the absence of a reliable experimental model has hampered our understanding of the role of the IECs in the pathogenesis of CD, we aimed to use the in vitro CD patient-derived organoids to perform preliminary functional studies.
Research plan and results obtained
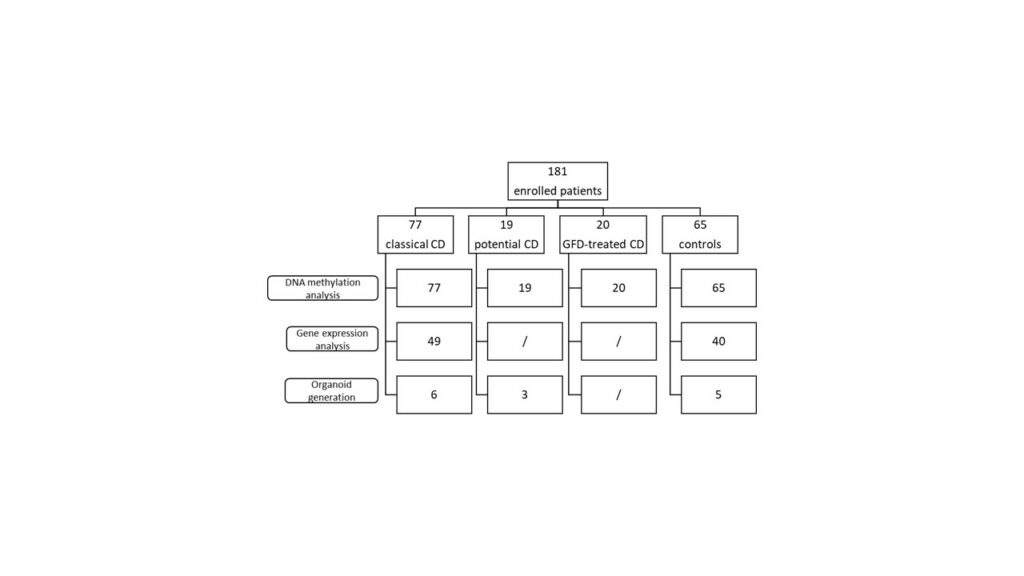
For this exploratory cohort study, a total of 181 children were enrolled: 77 classical CD, 19 potential CD, 20 GFD-treated CD, and 65 controls. Intestinal biopsies were obtained from all enrolled patients. DNA methylation and gene transcription analysis were performed on IECs to identify CD-specific epigenetic alterations (Figure 1).
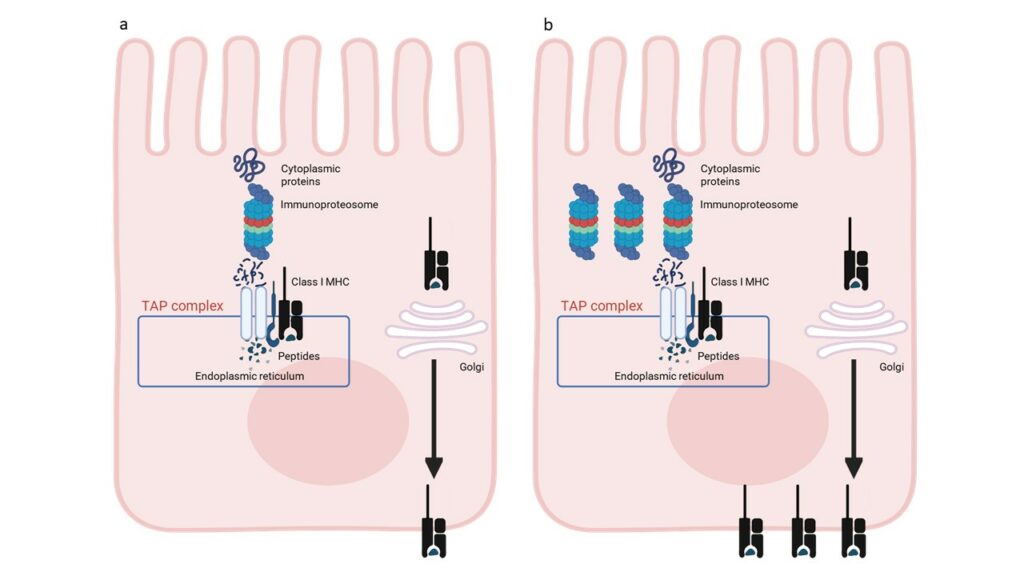
Differential methylation analysis identified 139 differentially methylated genes shared by each group of CD patients compared with controls. These data indicate that classical and potential CD patients share a common epigenetic profile at the IEC level, which is retained after GFD. Pathway analysis revealed that the DMGs are involved in 15 statistically significant pathways belonging to the antigen processing and presentation network. Gene expression data analysis revealed that transcriptional enriched patterns are also related to antigen processing and presentation pathways. These omics data indicate that IECs from CD patients are characterized by an enhanced antigen processing and presentation pathway (Figure 2).
To test whether the epigenetic signature of the original tissue is retained in CD-derived organoids, we generated intestinal organoids from a subset of children. DNA methylation analysis showed that CD-specific epigenetic changes were stable and maintained in the organoid model.
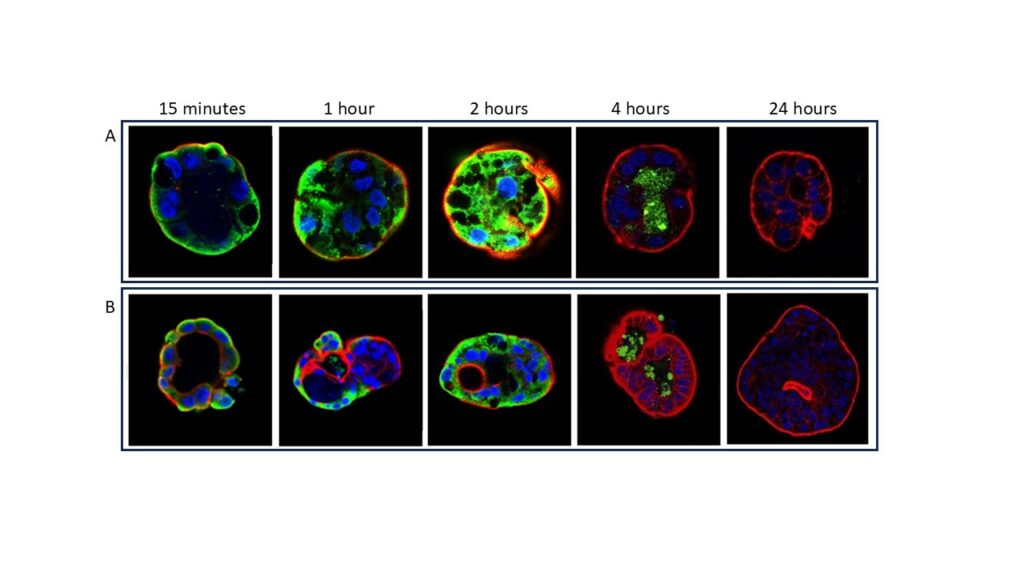
Organoids were then used to evaluate the interaction of organoid cells with a toxic gliadin peptide (P31-43) or with one of its scrambled forms. Confocal microscopic analysis revealed that P31-43 can specifically enter the cells. No interaction was detected after treatment with a scramble form of P31-43. The amount of internalized peptide seemed treatment-time dependent: the fluorescent signal is greater after 4h, at 8h the peptide is concentrated in the lumen of the organoids, and at 24h the signal is completely lost. Moreover, the peptide fluorescent signal appeared more intense in organoids from patients with CD (Figure 3).
Experimental design and methodologies
Consecutive children undergoing clinically indicated upper gastrointestinal endoscopy under deep sedation were enrolled for intestinal biopsy collection. Duodenal biopsy samples were freshly processed, and IECs were separated and purified via enzyme digestion and magnetic bead sorting for the Epithelial Cell Adhesion Molecule (EpCAM). DNA and RNA were extracted simultaneously. The genome-wide DNA methylation profile was obtained using the Illumina Infinium Human Methylation EPIC BeadChip array, and expression profiling was performed using RNA sequencing. All subsequent bioinformatic analyses were conducted using the R software. Organoids were generated from intestinal biopsies by isolating intestinal crypts. To evaluate the interaction of intestinal organoids with the toxic gliadin peptide P31-43, their polarity was inverted, and the apical-out organoids were treated with P31-43 (LGQQQPFPPQQPY) or one of its scrambled forms (YQPLFQPQGPQPQ), both tagged with a fluorescent dye.
Potential pitfalls and caveats
A limitation of our research is that our results are not yet supported by extensive functional studies. In particular, the involvement of antigen processing and presentation pathways in the pathogenesis of CD needs further investigation. However, our preliminary results in intestinal epithelial organoids indicate that this new tool can provide a reliable and previously unavailable model to investigate the role of the intestinal epithelial cells in the pathogenesis of CD.
Conclusions and discussion
This comprehensive study employed an omics profiling approach to investigate the involvement of the intestinal epithelium in the pathogenesis of celiac disease. We found CD-specific changes in IECs of children with CD regardless of the diet therapy. These data indicate that IECs from CD patients are characterized by an enhanced antigen processing and presentation pathway and may be actively involved in the pathogenesis of CD.
The CD-specific epigenetic changes in the intestinal epithelium are retained in the epithelium cultured ex-vivo as organoids. Therefore, the use of these patient-derived intestinal organoids might be very promising as a research tool for further investigating the epigenetic changes involved in the pathogenesis of CD. A preliminary evaluation of the interaction of toxic gliadin peptides with intestinal organoids revealed a specific interaction with P31-43 and suggested a higher affinity of organoids from CD patients. Further studies are needed to confirm this preliminary finding and to elucidate the mechanism of gliadin-organoid interaction.
