closed
Salvatore Oliva, 'La Sapienza' University, Rome, Italy
2-years Project
Celiac disease
Area: Clinics
Grant: 004/2020
- Title: Machine Learning As A New Method For “Case -Finding” In Celiac Disease (Needle Study).
- Duration: 2 Years Project
- Principal Investigator: Salvatore Oliva, La Sapienza University, Rome, Italy
Publications originating from the Project
- manuscript in preparation
THE STUDY
Project rationale and aims
Celiac disease (CeD) is a common autoimmune disorder affecting 1–2% of the global population. Despite well-defined diagnostic protocols, patients with atypical or nonspecific symptoms often remain undiagnosed. This delay in diagnosis can lead to serious health complications due to untreated disease. The NEEDLE project aimed to leverage artificial intelligence (AI), specifically Machine Learning (ML), to identify early indicators of CeD in children. By analyzing 73 clinical parameters—including symptoms, laboratory tests, and family history of autoimmune diseases—the study developed a model capable of predicting CeD with high accuracy. The ultimate goal was to support clinicians in improving early detection and optimizing diagnostic workflows.
Research plan and results obtained
The study included 325 children diagnosed with CeD and 490 age- and gender-matched controls. A supervised ML model, known as LASSO (Least Absolute Shrinkage and Selection Operator), was employed to analyze the dataset and identify relevant features predictive of CeD. The model consistently identified 40 significant features (Figure 1) across 20 training trials, achieving an average Area Under the Curve (AUC) of 0.77 (Figure 2). Key predictors included muscle pain, fatigue, gastroesophageal reflux symptoms, and family history of autoimmune diseases. This ML-based approach demonstrated its potential to improve case-finding strategies by focusing on nonspecific clinical presentations.
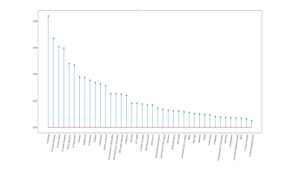
Figure 1: The 40 features identified by LASSO model.
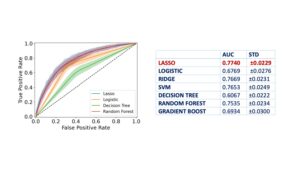
Figure 2: Performance in terms of Area Under the Curve (AUC) for the Receiver Operating Characteristic (ROC) curve for ML methods tested.
Experimental design and methodologies
A discovery cohort of children with CeD and matched controls was assembled, excluding data from specific antibody tests to better simulate real-world scenarios where these tests may not be immediately available. The LASSO model was trained and validated using cross-validation techniques to ensure robust predictions. The dataset was balanced through oversampling and undersampling techniques to mitigate class imbalances. Additional ML models, such as Support Vector Machines and Random Forest, were tested to compare performance.
Potential pitfalls and caveats
The primary limitation of this study lies in the model’s generalizability. As the training data was derived from a single geographic and demographic cohort, the results may not be fully applicable to other populations. Additionally, the exclusion of antibody test data, while purposeful, may limit the diagnostic accuracy of the model in settings where antibody tests are readily available. Further validation across diverse populations and broader age ranges is needed.
Conclusions and discussion
The NEEDLE project demonstrates the promising role of Machine Learning in improving early detection of celiac disease. The LASSO model effectively identified 40 predictive features, enabling clinicians to screen children with nonspecific symptoms more accurately. This approach could reduce the need for mass screening and focus resources on high-risk individuals. Future research should focus on validating this model in larger, more diverse cohorts and integrating it into clinical workflows to enhance diagnostic efficiency. By identifying subtle predictors, the NEEDLE project highlights the potential of AI in revolutionizing case-finding strategies for CeD, ultimately reducing undiagnosed cases and improving patient outcomes.
