closed
Giovanna Del Pozzo: Istituto di Genetica e Biofisica-CNR, Naples, Italy
Annual Project
Celiac disease
Area: Immunology
- Grant: FC 014/2014
- Title: Study of gene expression of HLA-DQ alleles associated to celiac disease in anti-gluten CD4+T cell immune response
- Topic: Genetic and Immunology of Celiac Disease
- Duration: Annual Project
- Principal Investigator: Giovanna Del Pozzo, CNR Naples (IGB-CNR)
- Partnerships: Carmen Gianfrani, Istituto di Biochimica delle Proteine del CNR (IBP-CNR)
Publications originated from the Project:
- Pisapia L, Camarca A, Picascia S, et al. HLA-DQ2.5 genes associated with celiac disease risk are preferentially expressed with respect to non-predisposing HLA genes: Implication for anti-gluten T cell response. J Autoimmun. 2016;70:63‐72. doi:10.1016/j.jaut.2016.03.016 https://pubmed.ncbi.nlm.nih.gov/27083396/
- Gianfrani C, Pisapia L, Picascia S, Strazzullo M, Del Pozzo G. Expression level of risk genes of MHC class II is a susceptibility factor for autoimmunity: New insights. J Autoimmun. 2018;89:1‐10. doi:10.1016/j.jaut.2017.12.016 https://pubmed.ncbi.nlm.nih.gov/29331322/
- Farina F, Picascia S, Pisapia L, et al. HLA-DQA1 and HLA-DQB1 Alleles, Conferring Susceptibility to Celiac Disease and Type 1 Diabetes, are More Expressed Than Non-Predisposing Alleles and are Coordinately Regulated. Cells. 2019;8(7):751. Published 2019 Jul 19. doi:10.3390/cells8070751 https://pubmed.ncbi.nlm.nih.gov/31331105/
- Pisapia L, Cerillo I, Farina F, et al. The HLA-DRB1 risk alleles for multiple sclerosis are differentially expressed in blood cells of patients from Southern Italy. Int J Immunogenet. 2019;46(6):479‐484. doi:10.1111/iji.12450 https://pubmed.ncbi.nlm.nih.gov/31313885/
- Pisapia L, Picascia S, Farina F, Barba P, Gianfrani C, Del Pozzo G. Differential expression of predisposing HLA-DQ2.5 alleles in DR5/DR7 celiac disease patients affects the pathological immune response to gluten. Sci Rep. 2020 Oct 14;10(1):17227. doi: 10.1038/s41598-020-73907-2. PMID: 33057065; PMCID: PMC7560598 https://pubmed.ncbi.nlm.nih.gov/33057065/
THE STUDY
Project rationale and aims
We have investigated the ability of professional Antigen Presenting Cells (APC) such as B lymphocytes (B-LCL) and dendritic cells (DC), carrying DQ2.5 genes either in homozygosis (DR3/DR3) or heterozygosis (DR1/DR3 and DR3/ DR5) to present gluten peptides to specific CD4 + T cells in relation to the mRNA and protein expression of each DQA1*05 and DQB1*02 gene. The hypothesis was that the level of stable gliadin peptide-DQ2.5 complexes on the APC surface, a key step to activate the inflammatory cascade in CD patients, may reflect not only the genetic configuration but also the effective DQA1*05 and DQB1*02 gene expression.
Figure 1
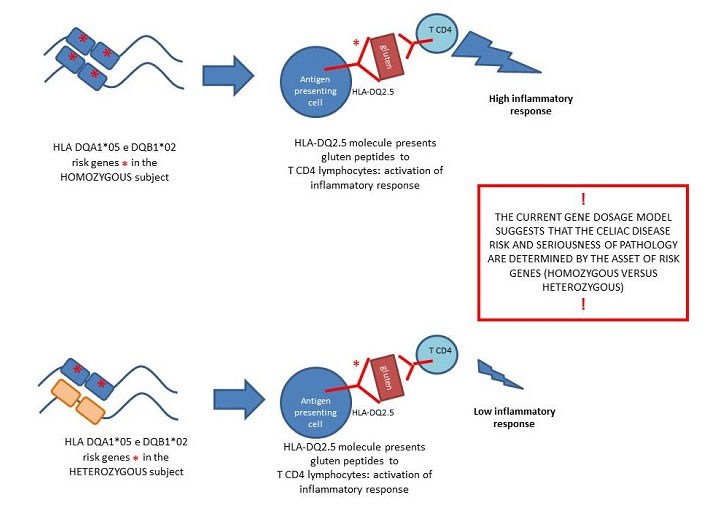
Figure 1 –The current gene dosage model suggests that the celiac disease risk and seriousness of pathology are determined by the HLA class II asset of risk genes ( homozygous versus heterozygous): this model does not explain some discrepancies found in the classification of patients into different risk classes. Our study aimed to clarify and complete the model of genetic risk by the measurements of the HLA-DQ2.5 proteins, on the surface of antigen presenting cells, and by the quantification of messengers DQA1*05 and DQB1*02 encoding the surface molecule. .
Research plan and results obtained
1) We measured the activation of gluten peptide-specific CD4+ T cells, using as APCs either DQ2.5 homozygous (DR3/DR3) and heterozygous (DR1/DR3 and DR3/DR5) B-LCLs or DCs, obtained from either celiac and healthy controls.
The results demonstrate that the activation threshold of an anti-gluten CD4+ T cell response in subjects with CD does not strictly correlate with the DQ2.5 genotype of professional APC. These experiments strongly suggested that the achievement of the activation threshold of pathogenic CD4+ T cells is primarily dependent on the concentration of gluten immunogenic peptides.
2) We assessed the effective surface amount of DQα1*05 and DQβ1*02 single chains by specific monoclonal antibodies in B-LCLs from homozygous and heterozygous patients. These results clearly demonstrated that similar capabilities of DQ2.5 homozygous and heterozygous APCs to activate anti-gluten CD4+ T cells are due to a comparable surface expression of DQα1*05 and DQβ1*02 proteins that form the DQ2.5 heterodimer.
3) We have measured the mRNA amounts of CD-associated DQA1*05 and DQB1*02 alleles and of HLA alleles not associated with the disease. In B-LCLs and DCs heterozygous for DQA1*05, the percentage of DQA1*05 mRNA was higher than the percentage of other DQA1*01 mRNA non-CD associated and not significantly different compared to that expressed by homozygous cells. Similar results were obtained for DQB1*02 mRNA which amount is greater compared to the expression of DQB1*03, *05 and *06 mRNA of heterozygous cells.
Experimental design and methodologies
For functional tests we used B-LCLs or DCs as APC to present specifigliadin peptides to gliadin-specific T cell lines and T cell clones. The CD4+T cells activation was assessed trough IFN-γ production by ELISA test and cell proliferation assay by [3H]thymdine incorporation.
Potential pitfalls and caveats
Conclusions and Discussion
A direct correlation between the genetic configuration of DQ2.5 haplotypes, the expected amount DQ2.5 heterodimer and the magnitude of T cell response to gluten has been demonstrated in previous studies. However, the “gene dosage theory”, does not fully explain the correlation of predisposition of CD among certain HLA genotypes and the pathogenic CD4+ T cell reactivity.
Here, we demonstrated that HLA DQA1*05 and DQB1*02 gene expression is much higher than expression of non-CD-associated genes. This influences the protein levels and causes a comparable cell surface exposure of DQ2.5 heterodimers between DQ2.5 homozygous and heterozygous celiac patients. As a consequence, the magnitude of the anti-gluten CD4+ T cell response is strictly dependent on the antigen dose and not on the DQ2.5 gene configuration of APCs. Furthermore, our findings support the concept that the expression of DQ2.5 genes is an important risk factor in celiac disease. The preferential expression of DQ2.5 alleles provides a new functional explanation of why these genes are so frequently associated with celiac disease and with other autoimmune disorders.
Figure 2
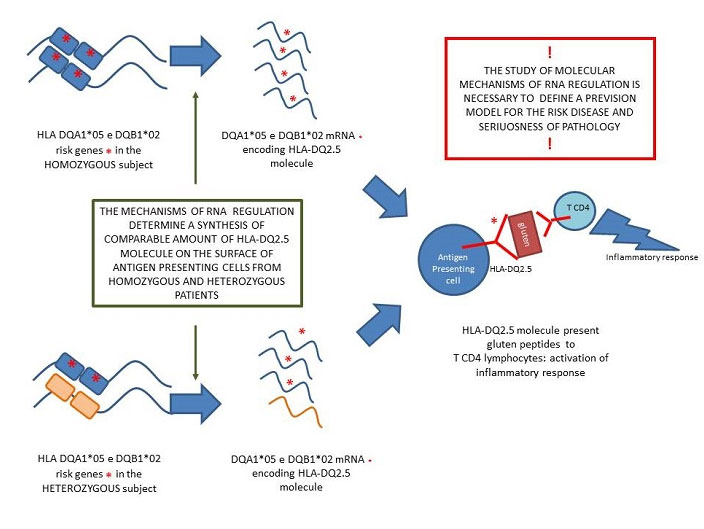
Figure 2 – The study demonstrated that the amount of HLA-DQ2.5 protein, presenting the gluten peptides, does not depend only on the homozygosity or heterozygosity of HLA risk genes, but also by the mRNA amount. Infact, when DQA1*05 and DQB1*02 genes are on the same chromosome produce more RNA than the corresponding alleles on the other chromosome: the result is a comparable amount of RNA produced by homozygous and heterozygous antigen presenting cells . These results complete the gene dosage model because and might support the appropriate classification of patients into risk classes.
