closed
Donatella Cielo, University Federico II, Naples, Italy
Triennial Fellowship
Celiac Disease
Area: Genetics
Tutor (Head Lab): Prof. Renata Auricchio, University Federico II, Naples
- Grant: FC 007/2017
- Title:LONG NON-CODING RNA EFFECTS ON THE DEVELOPMENT OF CELIAC DISEASE.
- Duration: Triennial Project
- Principal Investigator: Donatella Cielo, University Federico II, Naples
- Tutor (Head Lab): Prof. Renata Auricchio, University Federico II, Naples
Project rationale and aims
Non-coding-RNAs (ncRNAs), represent the majority of human transcripts and are involved in the post-transcriptional regulation of gene expression. Aim of this study is to evaluate the function and localization of lncRNAs putatively associated with Celiac Disease(CD).
We propose to evaluate the effect of 7 CD-associated SNPs, putatively affecting lncRNAs function, in keys CD cells types, observing directly their localization in formalin-fixed-paraffin-embedded(FFPE) sections of small-intestinal biopsies of celiac patients (CeD), controls (CTR), and potential celiac patients by RNAscope-assay technique, and quantifying their expression by Real-time-PCR in epithelial and non-epithelial sorted intestinal cells.
And finally, through a bioinformatics analysis, we explored the target genes and pathways regulated by these lncRNA.
Research plan and results obtained
In the first phase of study we analysed the expression of 7 lncRNA associated with CD in intestinal biopsies of 20 CeD and 13 CTR. Five of the LncRNA (AC007220.1, LINC01890, CCDC26, LINC02435, AL080284.1) were not found to be expressed at the intestinal level, while the LncRna AL139246 showed similar expression in both compartments (epithelium and lamina propria), without differences between CeD and CTR. Instead linc01882, which is associated with the PTPN2 locus, is mainly expressed in the lamina propria, respect to epithelium, both in CeD and in CTR. Inasmuch, this LncRNA is much more expressed in the celiac population respect to CTR
We analyzed, by RNAscope technology, the localization of linc01882 in FFPE of the samples analyzed in the molecular analysis and others selected by our biobank (10 CeD, 5 CTR, 4 potentials). Immunohistochemistry showed that linc01882 is present in the epithelium but mostly in lamina propria.
Subsequently the number of cases was increased and has been analysed the expression of linc01882 by RNAscope technology in 27 CeD, 10 potentials, 5 gluten-free diet patients (GFD) and 22 CTR. Linc01882 It is mainly expressed in the celiac population (figure 1A). In patients on gluten-free diet (GFD) the expression is similar to CeD, it decreases in potentials and CTR ( figure 1 B,C,D).
Then we performed the ISH in CeD with different degrees of mucosal atrophy, mild with partial villous atrophy, moderate with partial atrophy , severe with total atrophy. The expression of linc01882 in three groups is almost the same.
We perfomed Immunohistochemical staining for CD3 since from the previous experiments we hypothesized that linc01882 was probably expressed by the lymphocyte population. In figure 2 we showed the percent of T-cells present on the total of inflammatory cells in our population. As we can see from the graph, the percentage of T cells is higher in celiacs than in potential and controls. The figure 3 shows the linc01882-CD3 co-staining in CeD (dot red lncRna, dot brown CD3). In figure 4 we show that the expression of linc01882 (red line) correlates with the percentage of T-cell in the three groups.
By bioinformatics analysis we have identified the linc01882 target genes and the pathways regulated by it. This genes are involved in cellular processes such as inflammation and activation of T-cells, which have an important role in the pathogenesis of CD.
We analyzed, by Real-TimePCR the expression of some of this genes , ZEB1, MAP2K4 and PTPN2. PTPN2 and MAP2K4 genes show a lower level of expression in CeD than in CTR; ZEB1 shows the same levels of expression among the populations. MAP2K4 is a component of the kinase cascade that mediates cellular responses to cytokine signals and ZEB1 encodes a transcription factor that represses IL2 expression in T cells when it cooperates with the co-repressor C-terminal-binding protein (CtBP) . PTPN2 encodes PTPN2, also known as T cell PTP, and has been shown to act on several phosphorylated proteins, such as Janus-activated kinases (JAKs) and signal transducers and activators of transcription (STATs), to regulate cytokine signalling.
Furthermore, all the samples were genotyped for the PTPN2 SNP rs12971201, which is associated with the expression of linc01882. The “A” risk allele is most frequent in CeD . In the whole population, the “A” allele is less expressed. We evaluated if the genotype was associated to change in linc01882-target gene expression. There is no association with any of genes analyzed, confirming the literature data according which the SNP is not a cis-eQTL.
Figures
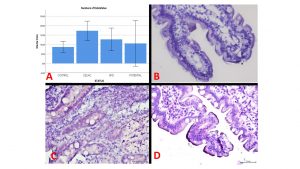
Figure 1. The average value of signals (dots) in 100 cells considering 3 different portions of each biopsy(A).The expression by ISH of linc01882 in CTR (B), CeD (C) and potential (D)
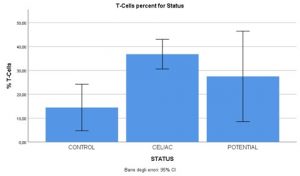
Figure 2. The percent of T-cells present on the total of inflammatory cells in our population.
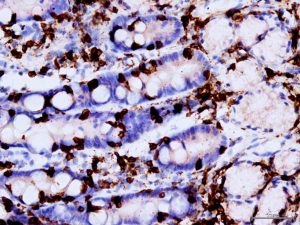
Figure 3. linc01882-CD3 co-staining in CeD patient (dot red lncRna, dot brown CD3.
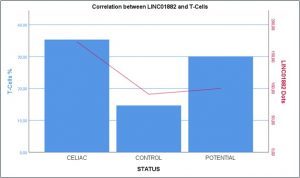
Figure 4.The expression of linc01882 (red line) correlates with the percentage of T-cell in the three groups(CeD,CTR,Potentials).
Experimental design and methodologies
Extractions RNA and DNAThe RNeasy FFPE Kit and Dnasy FFPE Kit were used for RNA and DNA extractions from formalin-fixed, paraffin-embedded intestinal biopsies sections of CeD, CTR, GFD and potential.
Real-time PCR was performed by TaqMan methodology (7900T) using TaqMan GeneExpression (Life Technologies) probes. The relative expression of each gene was obtained by using the ΔΔct method and normalized to the expression of an endogenous gene GUSb. The genes analysed are MAP2K4, ZEB1 and PTPN2.The custom TaqMan Gene Expression assays were provided by Thermofischer
Genotyping.The SNP(rs12971201) was genotyped using TaqMan technology (Life Technologies) by TaqMan SNP Genotyping Assays on a 7900HT Fast Real-Time PCR System (Applied Bio systems).
BaseScope in Situ Hybridization(ISH).Starting with properly prepared tissue samples ,sections mounted on glass slides are first pretreated, and then RNA-specific probes are hybridized to target RNA. The signal is amplified using a multi-step process, and detected using a red-chromogenic substrate. Each single RNA transcript appears as a distinct dot of chromogen precipitate visible using a common bright field microscope at 40–100X magnification.Single-molecule signals can be quantified on a cell-by-cell basis by manual counting
Immunohistochemical staining for CD3.CD3 (2GV6, VENTANA) monoclonal antibody is intended to be used for the qualitative identification by light microscopy of human CD3 molecule in formalin-fixed, paraffin-embedded tissue by immunohistochemical staining using the automated BOND system(LEICA).
Statistical analyses were performed with SPSS 17.0 (SPSS Inc., Chicago, IL, USA).
Potential pitfalls and caveats
The difficulties and limitations encountered are mainly linked to the pandemic due to the Sars Covid-19 virus. Unfortunately,It was not possible to develop the last part of the project, which involved the development of organoids from intestinal cells from fresh intestinal biopsies. For this purpose it was necessary to recruit patients through the DH of gastroenterology of the Department of Pediatrics which remained closed for a long time. In addition, I encountered other problems due to delays in delivering the laboratory material needed to complete the experiments.
Conclusions and discussion
We focused on molecular biology and immunohistochemistry studies, which gave promising results in the second year. First of all we aimed at expanding the number of cases to be analyzed; we performed further gene expression and genotyping studies that allowed us to explore the genes and pathways regulated by lncRna (linc01882). By immunohistochemistry we wanted to characterize the cells that expresses this lncRna and in CeD to evaluate an eventual correlation of the degree of mucosal atrophy with the expression of linc01882.
The study suggests, that linc01882 plays a role in the development of CD, in association with rs12971201 PTPN2-SNP. The expression of lncRNA is not affected by the degree of inflammation and is stable also during gluten free diet.
The results prove that, there is no more difference in the expression between the two compartments, epithelium and lamina propria in comparison with the preliminary data. In fact, increasing the number of cases we noticed that in some CeD the expression of linc01882 is very high also in the epithelium.We have hypothesized that the signals we saw in the epithelium were due to the presence of intaepithelial lymphocytes, so we considered appropriate to perform the co-staining with anti-CD3 to demonstrate that it was due to lymphocyte and therefore the lncRna was mainly expressed by the T-cells.
The data show that the expression of linc01882 correlates with the presence of T-cells, so it is clear that lncRNA is expressed by the lymphocyte population. Furthermore, there is no difference in expression in the various degrees of mucosal atrophy in the celiac population, suggesting that the expression of lncRna is not affected by the degree of inflammation. This hypothesis is strengthened by the fact that in GFD patients the expression of linc01882 is comparable to the levels of expression found in active CeD.
The main finding in our study is that genetic variations in the PTPN2 locus are involved in the regulation of the lncRNA linc01882 that is mainly expressed in T cells. These cells play an important role in controlling autoimmune responses.
The study suggests, for the first time, that linc01882 plays a role in the development of CD, in association with rs12971201 PTPN2-SNPs. Further detailed studies would be necessary to assess the exact role of linc01882 in T-cells in relation to disease development.
